Nat Med | ʻO kahi ala multi-omics i ka palapala ʻana i ka ʻōpū i hoʻohui ʻia, immune a me ka ʻāina microbial o ka maʻi maʻi colorectal e hōʻike ana i ka pilina o ka microbiome me ka ʻōnaehana pale.
ʻOiai ua aʻo nui ʻia nā biomarkers no ka maʻi maʻi kolona mua i nā makahiki i hala iho nei, ua hilinaʻi nā alakaʻi lapaʻau o kēia manawa i ka hana ʻana o ka tumor-lymph node-metastasis staging a me ka ʻike ʻana i nā hemahema DNA mismatch repair (MMR) a i ʻole microsatellite instability (MSI) (me ka hoʻāʻo ʻana i nā pathology maʻamau) e hoʻoholo ai i nā ʻōlelo lapaʻau. Ua ʻike ka poʻe noiʻi i ka nele o ka hui ʻana ma waena o nā pane kūlohelohe e pili ana i ka hōʻike ʻana o ke ʻano, microbial profiles, a me ka stroma tumor i loko o ka Cancer Genome Atlas (TCGA) colorectal cancer cohort a me ke ola maʻi.
Ke holomua nei ka noiʻi ʻana, ua hōʻike ʻia nā hiʻohiʻona nui o ka maʻi kanesa colorectal mua, e pili ana i ka maʻi maʻi cellular, immune, stromal, a i ʻole microbial ʻano o ka maʻi maʻi, ua hōʻike ʻia e pili nui me nā hopena lapaʻau, akā aia nō ka palena o ka hoʻomaopopo ʻana i ka hopena o kā lākou mau pilina i nā hopena maʻi.
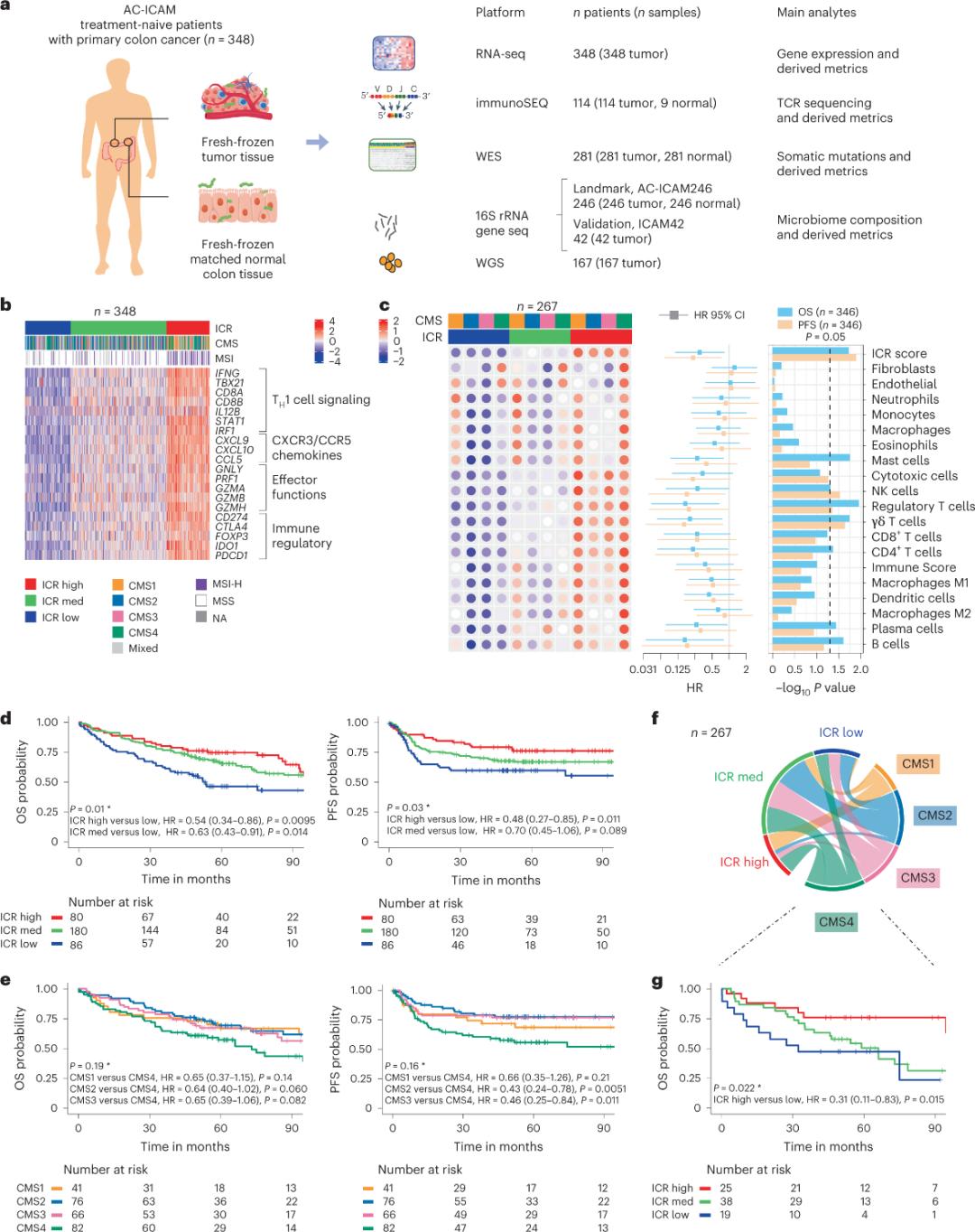
No ka wehewehe ʻana i ka pilina ma waena o ka paʻakikī phenotypic a me ka hopena, ua hoʻomohala a hoʻopaʻa ʻia kahi hui o nā mea noiʻi mai ka Sidra Institute of Medical Research ma Qatar i kahi helu i hoʻohui ʻia (mICRoScore) e hōʻike ana i kahi hui o nā maʻi me nā helu ola maikaʻi ma o ka hoʻohui ʻana i nā ʻano microbiome a me ka immune rejection constants (ICR). Ua hana ka hui i kahi loiloi genomic piha o nā laʻana hau hau hou mai 348 mau mea maʻi me ka maʻi maʻi colorectal mua, e komo pū ana me ka RNA sequencing o nā maʻi maʻi a me ke ʻano o ka ʻiʻo colorectal maikaʻi, ke kaʻina exome holoʻokoʻa, ka hohonu T-cell receptor a me ka 16S bacterial rRNA gene sequencing, i hoʻohui ʻia e ka holoʻokoʻa genome sequencing e hōʻike hou i ka microbiome. Ua paʻi ʻia ke aʻo ʻana ma Nature Medicine e like me "He tumo i hoʻohui ʻia, immune a me microbiome atlas o ka maʻi maʻi ʻaʻai koko".

ʻatikala i paʻi ʻia ma Nature Medicine
AC-ICAM Nānā
Ua hoʻohana ka poʻe noiʻi i kahi kahua genomic orthogonal no ka nānā ʻana i nā laʻana tumo maloʻo hou a hoʻohālikelike ʻia i ka ʻiʻo koko olakino e pili ana (ʻo ka maʻi maʻamau) mai nā mea maʻi me kahi hōʻailona histologic o ka maʻi maʻi ʻaʻai ʻole me ka ʻōnaehana systemic. Ma muli o ke kaʻina holoʻokoʻa holoʻokoʻa (WES), ka mana o ka maikaʻi o ka ʻikepili RNA-seq, a me ka nānā ʻana i nā koina hoʻopili, mālama ʻia ka ʻikepili genomic mai nā maʻi 348 a hoʻohana ʻia no ka nānā ʻana i lalo me kahi median follow-up o 4.6 makahiki. Ua kapa ka hui noiʻi i kēia kumu waiwai ʻo Sidra-LUMC AC-ICAM: He palapala ʻāina a me ke alakaʻi i nā pilina immune-cancer-microbiome (Figure 1).
Hoʻokaʻawale molekala me ka hoʻohana ʻana i ka ICR
ʻO ka hopu ʻana i kahi pūʻulu modular o nā hōʻailona genetic immune no ka immunosurveillance maʻi kanesa mau, i kapa ʻia ʻo ka immune constant of rejection (ICR), ua hoʻopaʻa ʻia ka hui noiʻi i ka ICR ma o ka hoʻohui ʻana iā ia i loko o kahi papa 20-gene e uhi ana i nā ʻano maʻi maʻi like ʻole, me ka melanoma, ka maʻi maʻi ʻaʻai, a me ka maʻi maʻi umauma. Ua pili pū ʻia ʻo ICR me ka pane immunotherapy i nā ʻano maʻi maʻi like ʻole, me ka maʻi maʻi umauma.
ʻO ka mea mua, ua hōʻoia ka poʻe noiʻi i ka hōʻailona ICR o ka hui AC-ICAM, me ka hoʻohanaʻana i ka ICR gene-based co-classification approach no ka hoʻokaʻawaleʻana i ka pūʻulu i loko oʻekolu mau puʻupuʻu / immune subtypes: ICR kiʻekiʻe (nā maʻi wela), ICR medium a me ICR haʻahaʻa (nā maʻi anuanu) (Figure 1b). Ua ʻike ka poʻe noiʻi i ka propensity immune e pili ana me ka consensus molecular subtypes (CMS), kahi helu transcriptome-based o ka maʻi maʻi kolona. ʻO nā māhele CMS i komo i CMS1/immune, CMS2/canonical, CMS3/metabolic a me CMS4/mesenchymal. Ua hōʻike ʻia ka hōʻike ʻana ua hoʻopili maikaʻi ʻia nā helu ICR me kekahi mau ala maʻi maʻi maʻi ma nā ʻano CMS āpau, a ua ʻike ʻia nā pilina maikaʻi me nā ala immunosuppressive a me ka stromal i nā maʻi maʻi CMS4 wale nō.
Ma nā CMS a pau, ʻoi aku ka kiʻekiʻe o ka mea pepehi maoli (NK) cell a me T cell subsets i ka ICR high immune subtypes, me ka ʻoi aku ka nui o nā ʻano leukocyte ʻē aʻe (Figure 1c).
Kiʻi 1. ʻO ka hoʻolālā haʻawina AC-ICAM, pūlima gene pili i ka immune, immune a me nā subtypes molecular a me ke ola.
Lawe ʻo ICR i nā pūnae T i hoʻonui ʻia i ka tumo
ʻO kahi liʻiliʻi wale nō o nā pūnae T e komo ana i ka ʻiʻo tumo i hōʻike ʻia he kikoʻī no nā antigens tumo (emi ma lalo o 10%). No laila, ʻo ka hapa nui o nā pūnaewele T intra-tumor i kapa ʻia ʻo ia nā pūnana T bystander (bystander T cell). ʻO ka pilina ikaika loa me ka helu o nā pūnaewele T maʻamau me nā TCR huahua i ʻike ʻia i loko o ka stromal cell a me ka leukocyte subpopulations (ʻike ʻia e RNA-seq), hiki ke hoʻohana ʻia e koho i nā subpopulations cell T (Figure 2a). I loko o nā pūʻulu ICR (ka holoʻokoʻa a me ka CMS classification), ua ʻike ʻia ka clonality kiʻekiʻe o ka immune SEQ TCRs i ka ICR-kiʻekiʻe a me CMS subtype CMS1 / mau pūʻulu pale (Figure 2c), me ka hapa kiʻekiʻe o nā maʻi koko ICR-kiʻekiʻe. Ke hoʻohana nei i ka transcriptome holoʻokoʻa (18,270 genes), ʻeono ICR genes (IFNG, STAT1, IRF1, CCL5, GZMA, a me CXCL10) i waena o nā ʻano he ʻumi kiʻekiʻe e pili ana me ka TCR immune SEQ clonality (Figure 2d). ʻOi aku ka ikaika o ka ImmunoSEQ TCR clonality me ka hapa nui o nā genes ICR ma mua o nā correlations i ʻike ʻia me ka hoʻohana ʻana i nā hōʻailona CD8+ pane-tumu (Figure 2f a me 2g). I ka hopena, hōʻike ka loiloi ma luna nei e hopu ka pūlima ICR i ka hele ʻana o nā pūnae T i hoʻonui ʻia me ka clonally a hiki ke wehewehe i nā hopena prognostic.
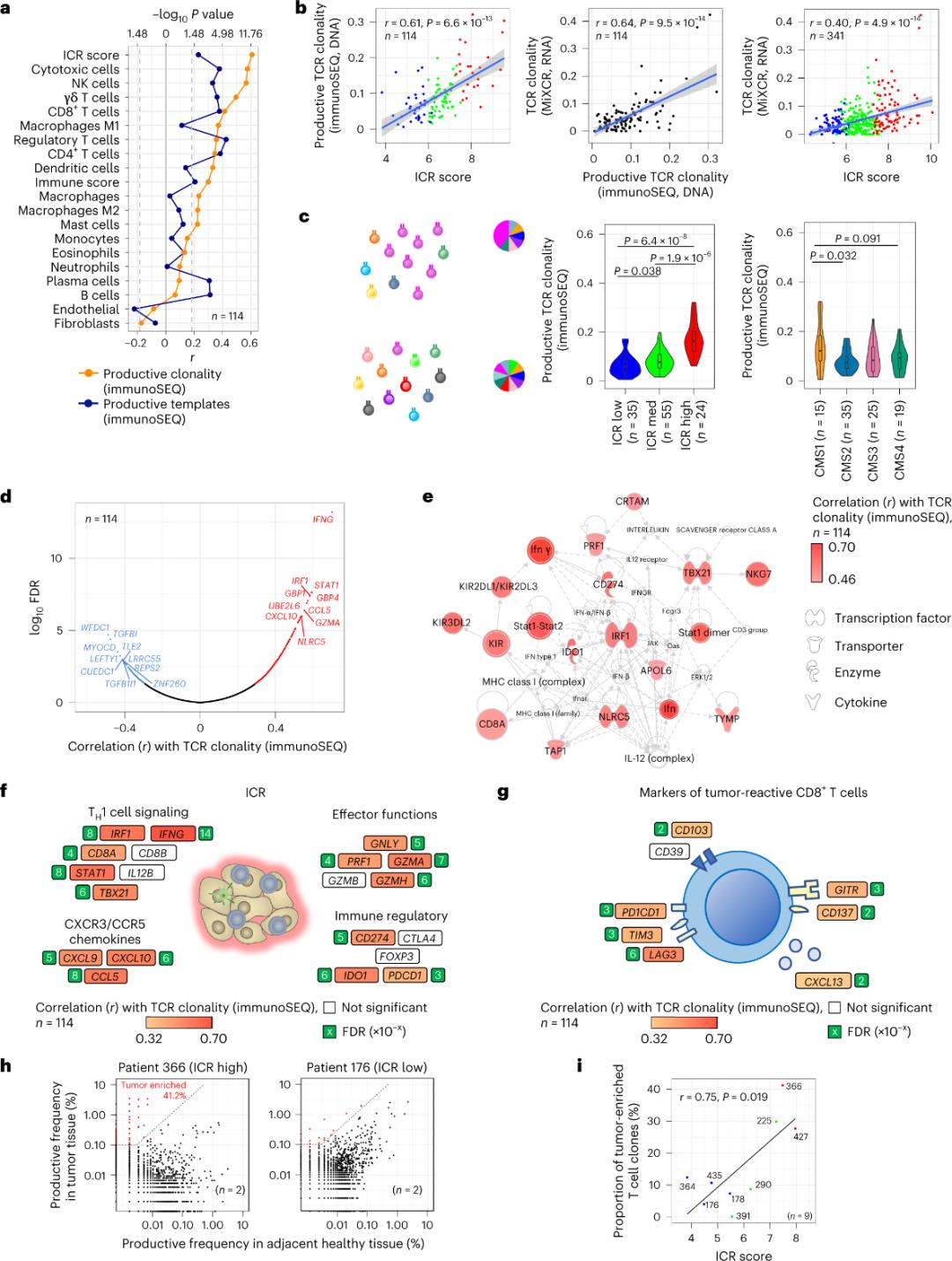
Kiʻi 2. TCR metric a me ka pilina me nā genes pili i ka immune, immune a me nā subtypes molecular.
Hoʻohui ʻia ka Microbiome i nā ʻiʻo maʻi maʻi olakino a me ka colon
Ua hana nā mea noiʻi i ka 16S rRNA sequencing me ka hoʻohana ʻana i ka DNA i unuhi ʻia mai ka maʻi maʻi like ʻole a me ka ʻiʻo koko olakino mai 246 mau maʻi (Figure 3a). No ka hōʻoia ʻana, ua ʻimi hou ka poʻe noiʻi i ka 16S rRNA gene sequencing data mai kahi 42 mau hōʻailona maʻi maʻi ʻaʻole i kūlike i ka DNA maʻamau i loaʻa no ka nānā ʻana. ʻO ka mea mua, ua hoʻohālikelike ka poʻe noiʻi i ka nui o ka flora ma waena o nā maʻi maʻi like ʻole a me ka ʻiʻo koko olakino. Ua hoʻonui nui ʻia ʻo Clostridium perfringens i loko o nā maʻi maʻi i hoʻohālikelike ʻia i nā laʻana olakino (Figure 3a-3d). ʻAʻohe ʻokoʻa koʻikoʻi o ka alpha diversity (ka ʻokoʻa a me ka nui o nā ʻano i loko o kahi laʻana hoʻokahi) ma waena o nā ʻōpū a me nā laʻana olakino, a ua ʻike ʻia kahi hoʻohaʻahaʻa haʻahaʻa o ka ʻokoʻa microbial i nā ʻōpū kiʻekiʻe ICR e pili ana i nā maʻi maʻi ICR-haʻahaʻa.
No ka ʻike ʻana i nā hui pili pili kino ma waena o nā profile microbial a me nā hopena lapaʻau, ua manaʻo nā mea noiʻi e hoʻohana i ka 16S rRNA gene sequencing data e ʻike i nā hiʻohiʻona microbiome e wānana i ke ola. Ma AC-ICAM246, ua holo nā mea noiʻi i kahi hiʻohiʻona regression OS Cox i koho i nā hiʻohiʻona 41 me nā coefficients non-zero (pili ʻia me ka ʻokoʻa o ka make make), i kapa ʻia ʻo MBR classifiers (Figure 3f).
I loko o kēia pūʻulu hoʻomaʻamaʻa (ICAM246), kahi helu MBR haʻahaʻa (MBR<0, MBR haʻahaʻa) i pili me ka haʻahaʻa haʻahaʻa o ka make (85%). Ua hōʻoia ka poʻe noiʻi i ka hui ma waena o ka MBR haʻahaʻa (pilikia) a me ka OS lōʻihi i ʻelua mau pūʻulu hoʻopaʻa kūʻokoʻa (ICAM42 a me TCGA-COAD). (Figure 3) Ua hōʻike ka haʻawina i kahi pilina ikaika ma waena o ka endogastric cocci a me nā helu MBR, i like me ka ʻōpū a me ka ʻiʻo koko olakino.
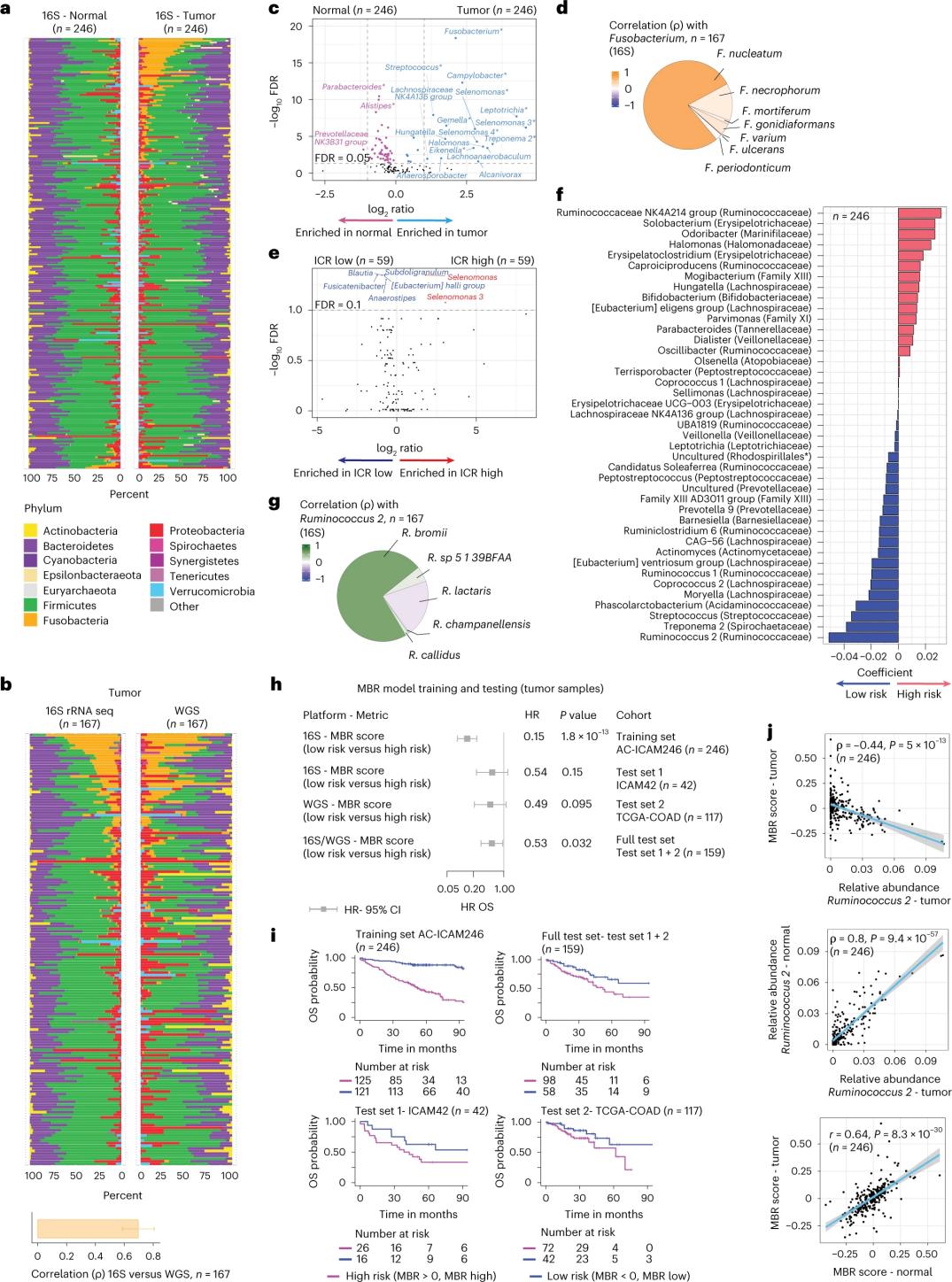
Kiʻi 3. ʻO Microbiome i loko o ka ʻōpū a me nā ʻiʻo olakino a me ka pilina me ka ICR a me ke ola maʻi.
Ka hopena
ʻO ke ala multi-omics i hoʻohana ʻia i kēia haʻawina e hiki ai ke ʻike pono a me ka nānā ʻana i ka pūlima molekala o ka pane ʻana i ka pale ʻana i ka maʻi maʻi colorectal a hōʻike i ka pilina ma waena o ka microbiome a me ka ʻōnaehana pale. Ua hōʻike ʻia ke kaʻina o ka TCR hohonu o nā ʻiʻo a me nā ʻiʻo olakino e pili ana ka hopena prognostic o ICR ma muli o kona hiki ke hopu i nā clones T cell i hoʻonui ʻia i ka tumo a me nā ʻāpana antigen-specific T.
Ma ka nānā ʻana i ka haku mele microbiome tumor me ka hoʻohana ʻana i ka 16S rRNA gene sequencing i nā laʻana AC-ICAM, ua ʻike ka hui i kahi pūlima microbiome (MBR risk score) me ka waiwai prognostic ikaika. ʻOiai ua loaʻa kēia pūlima mai nā laʻana tumo, aia kahi pilina paʻa ma waena o ka colorectum olakino a me ka helu MBR maʻi tumora, e hōʻike ana e hiki i kēia pūlima ke hopu i ka ʻōpū microbiome o nā maʻi. Ma ka hoʻohui ʻana i nā helu ICR a me MBR, ua hiki ke ʻike a hōʻoia i kahi biomarker haumāna multi-omic e wānana ana i ke ola ʻana o nā maʻi me ka maʻi maʻi ʻaʻai. Hāʻawi ka ʻikepili multi-omic o ka haʻawina i kumu e hoʻomaopopo maikaʻi ai i ka biology maʻi maʻi colon a kōkua i ka ʻike ʻana i nā ala lapaʻau pilikino.
Ka manawa hoʻouna: Iune-15-2023
 中文网站
中文网站